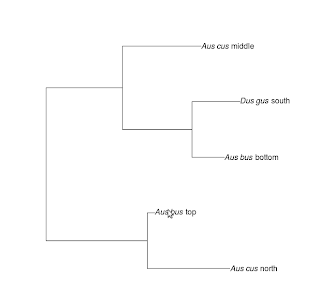
The following is a simple R-based solution for changing the tip labels of phylogenetic trees. First, we need to create a tree and a dataframe containing both the specimen codes and the ultimate labels.
library(ape)
tr <- rtree(5)
d1 <- c("t1","t2","t3","t4","t5")
d2 <- c( "paste(italic('Aus bus'), ' top')", "paste(italic('Aus bus'), ' bottom')", "paste(italic('Aus cus'), ' middle')", "paste(italic('Aus cus'), ' north')", "paste(italic('Dus gus'), ' south')" )
d <- as.data.frame(cbind(label=d1, nlabel=d2))
The code in the nlabel column contains code defining a plottable expression that enables scientific names to be formatted as italics. In my work, I saved this table as a separate file which I call with read.table("file.txt", header=TRUE, sep="\t", stringsAsFactors=FALSE, quote=""). The quote argument is important as it carries the nested quotes through into the dataframe properly.
The business of actually changing the tip labels is done with the following lines:
tr$tip.label<-d[[2]][match(tr$tip.label, d[[1]])]
tr$tip.label<-sapply(tr$tip.label, function(x) parse(text=x))
The first line enters the expressions for the new labels in the correct order. The second line converts the character string into a printable expression.
Plot the tree and voila!
2 comments:
You need to make the tip labels into character vectors before doing sapply. Otherwise, they all become the levels of a factor:
tr$tip.label<-as.character(d[[2]][match(tr$tip.label, d[[1]])])
tr$tip.label<-sapply(tr$tip.label, function(x) parse(text=x))
Hello, I could not plot the tree with your example.
Post a Comment